Active Grants
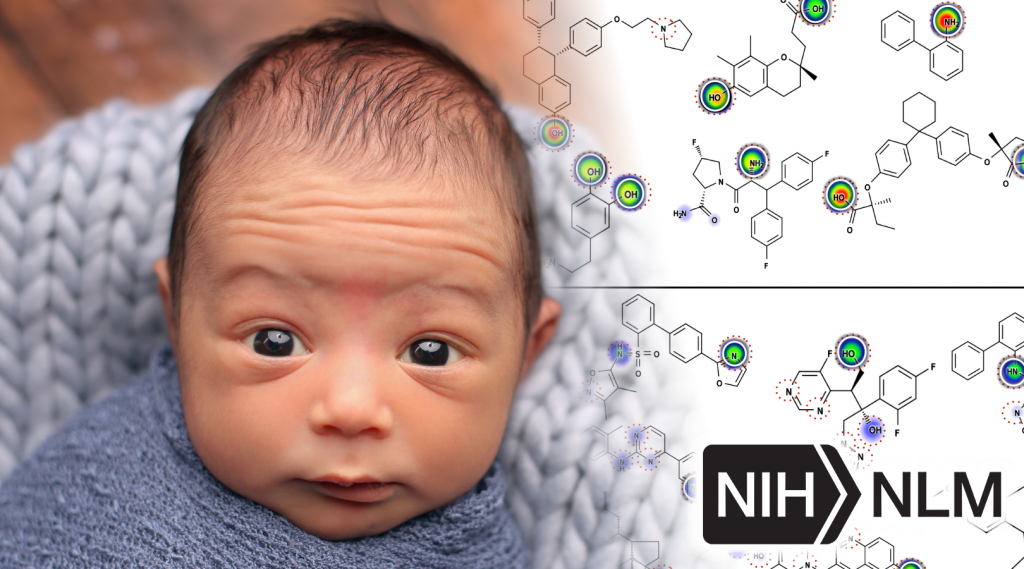
Currently we are funded by the NIH through the National Library Of Medicine to study Metabolism and Reactivity under award number R01LM012222, and Metabolism in Children under award number R01LM012482.
Summary of Work
My early work, during my PhD years in Pierre Baldi’s lab at UC Irvine, focused on applying machine learning to predict the biological properties of small molecules and improving computational algorithms for measuring structural similarity between molecules using fingerprints. This work includes a highly cited study that (1) is among the first studies to use support vector machines (SVM) to predict biological properties of small molecules and (2) is one of the early papers that sparked a resurgence of chemical informatics within the bioinformatics community. Later studies substantially improved on SVMs by using neural networks. I also discovered several seminal fingerprint search algorithms, including the first sub-linear search algorithm for fingerprints, making the first algorithmic progress in decades in this critically important area. Also relevant to this study, I was a key developer for one of the first public databases of commercially available chemicals (the ChemDB, http://cdb.ics.uci.edu/), which was one of a group of databases (including ZINC and ChemBank) that supported a renewed interest in chemical informatics amongst computational biologists and ultimately led the creation of PubChem.
- Swamidass, S. J., Chen, J., Bruand, J., Phung, P., Ralaivola, L., and Baldi, P. (2005) Kernels for small molecules and the prediction of mutagenicity, toxicity and anti-cancer activity. Bioinformatics, 21 Suppl 1:i359–368, Jun. PMID: 15961479
- Swamidass, S. J., Azencott, C. A., Lin, T. W., Gramajo, H., Tsai, S. C., and Baldi, P. (2009) Influence Relevance Voting: an accurate and interpretable virtual high throughput screening method. Journal of Chemical Information and Modeling, 49:756–766, Apr. PMCID: PMC2750043
- Swamidass, S. J. and Baldi, P. (2007). Bounds and algorithms for fast exact searches of chemical fingerprints in linear and sublinear time. J. Chem. Inf. Model, 47(2):302–317. PMCID: PMC2527184
- Chen, J., Swamidass, S. J., Dou, Y., Bruand, J., and Baldi, P. (2005). Chemdb: a public database of small molecules and related chemoinformatics resources. Bioinformatics, 21(22):4133. PMID: 16174682
In the first year as a resident at Washington University in St. Louis and as a visiting scientist at the Broad Institute of Harvard/MIT, I worked closely with the Broad’s high throughput screening (HTS) team to use economic modeling to improve the efficiency of expensive and time-consuming HTS experiments. This was a highly innovative approach that had never been attempted before and it proved extremely successful. Explicitly encoding costs and utility in an economic framework we designed led to large efficiency gains, sometimes tripling the rate at which active scaffolds were discovered from HTS screens. Some of this software is now publicly available and maintained by my group (https://bitbucket.org/swamidass/scaffold-network-generator/). This effort highlights a hallmark my current work: close collaboration with experimental scientists to produce useful tools that enable more efficient discovery.
- Swamidass, S. J., Bittker, J. A., Bodycombe, N. E., Ryder, S. P., and Clemons, P. A. (2010). An Economic Framework to Prioritize Confirmatory Tests after a High-Throughput Screen. Journal of Biomolecular Screening, 15(6):680. PMCID: PMC3069998
- Swamidass, S. J., Calhoun, B. T., Bittker, J. A., Bodycombe, N. E., and Clemons, P. A. (2011). Enhancing the rate of scaffold discovery with diversity-oriented prioritization. Bioinformatics, 27(16):2271–2278. PMCID: PMC3150035
- Swamidass, S. J., Calhoun, B. T., Bittker, J. A., Bodycombe, N. E., and Clemons, P. A. (2011). Utility-aware screening with clique-oriented prioritization. Journal of chemical information and modeling, 52(1):29–37. PMCID: PMC3264765
- Matlock, M. K., Zaretzki, J. M., and Swamidass, S. J. (2013) Scaffold network generator: a tool for mining molecular structures. Bioinformatics, 29(20):2655–2656. PMID: 23918250
My work on HTS screening continued into my first years on faculty at Washington University in St. Louis. Funded first by Pfizer and then by GlaxoSmithKline, I designed and implemented software systems that used historic HTS data to direct internal drug repurposing efforts. The exact drug development projects on which we used this software remain confidential, but this effort led to several publications in leading journals focused on the technical details of our approach. These publications include a body of work focused on how to interpret ambiguous or missing data in HTS experiments.
- Swamidass, S. J. (2011). Mining small-molecule screens to repurpose drugs. Briefings in Bioinformatics, 12(4):327–335. PMID: 2171546.
- Swamidass, S. J., Schillebeeckx, C. N., Matlock, M., Hurle, M. R., and Agarwal, P. (2014). Combined analysis of phenotypic and target-based screening in assay networks. Journal of Biomolecular screening, pages 782–790. PMID: 2456342.
- Calhoun, B. T., Browning, M. R., Chen, B. R., Bittker, J. A., and Swamidass, S. J. (2012). Automatically detecting workflows in PubChem. Journal of Biomolecular Screening, 17(8):1071–1079. PMID: 22693105.
- Browning, M. R., Calhoun, B. T., and Swamidass, S. J. (2013). Managing missing measurements in small-molecule screens. Journal of Computer-Aided Molecular Design, pages 1–10. PMID: 23585219.
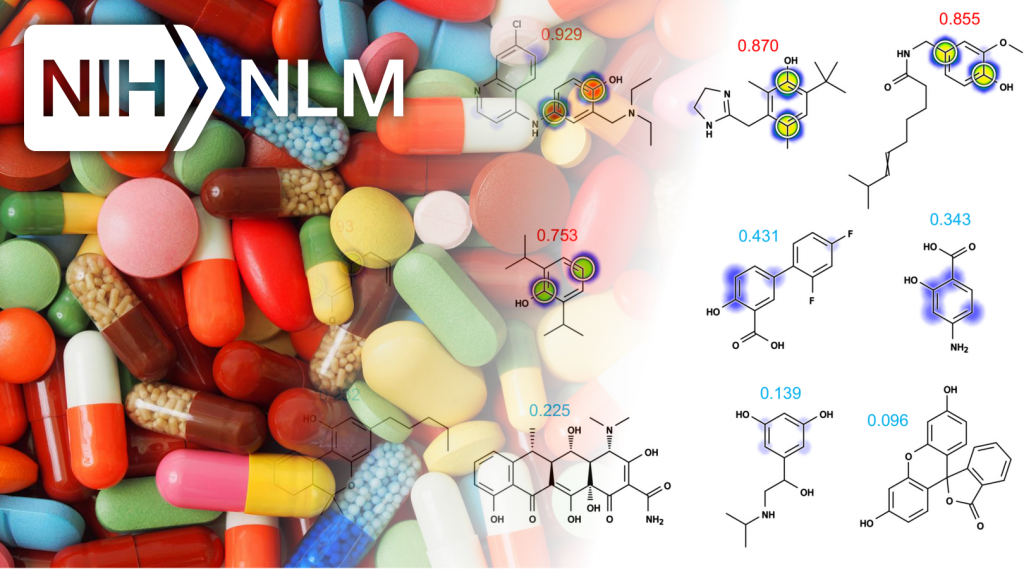
Most recently, my work has focused on computationally modeling metabolism and reactivity. This work has included improved accuracy of P450 metabolism predictions using neural networks, and the release of two web-available P450 prediction services based on RS-Predictor from Curt Breneman’s group at RPI (http://reccr.chem.rpi.edu/Software/RS-WebPredictor/) and XenoSite metabolism predictor from my group (https://swami.wustl.edu/xenosite). Just this year, my group demonstrated a particularly exciting result (in Bioinformatics) that shows we can accurately build P450 metabolism models from ambiguous, region-level training data. This result is a major advance forward in the field, and enables our methods to learn from data where the exact site of metabolism is not known. Recently, we have successfully extended this work to predict UGT metabolism (in preparation), reactivity with glutathione (in collaboration with Dr. Grover Miller and just accepted), and epoxidation (in preparation). This body of work demonstrates (1) a track record of steady improvement to existing approaches, (2) development of innovative approaches to incorporate, until now, unused data, (3) a commitment to disseminate our work in useful tools, and (4) the direct extension of these approaches beyond P450 sites of metabolism to other critical metabolic and reactivity pathways. The current proposal builds logically on this work into an area of critical clinical importance: the curation of data and development of tools to support reactive metabolite modeling.
- Zaretzki, J., Matlock, M., and Swamidass, S. J. (2013). XenoSite: Accurately predicting CYP-mediated sites of metabolism with neural networks. Journal of Chemical Information and Modeling, 53(12):3373–3383. PMID: 24224933
- Zaretzki, J., Bergeron, C., Huang, T., Rydberg, P., Swamidass, S. J., and Breneman, C. (2012). RS-WebPredictor: A server for predicting CYP-mediated sites of metabolism on drug-like molecules. Bioinformatics. PMCID: PMC3570214
- Matlock, M. K., Hughes, T. B., and Swamidass, S. J. (2014). XenoSite-server: A web-available site of metabolism prediction tool. Bioinformatics,2015. PMID: 25411327
- Zaretzki, J., Browning, M. R., Hughes, T. B., and Swamidass, S. J. (2015) Extending P450 Site-of-Metabolism Models with Region-Resolution Data. Bioinformatics 2015.

You must log in to post a comment.