Deep Learning to Predict the Formation of Quinone Species in Drug Metabolism
Hughes, T. B. and Swamidass, S. J. (2017). Chemical Research in Toxicology, 30(2), 642–656.
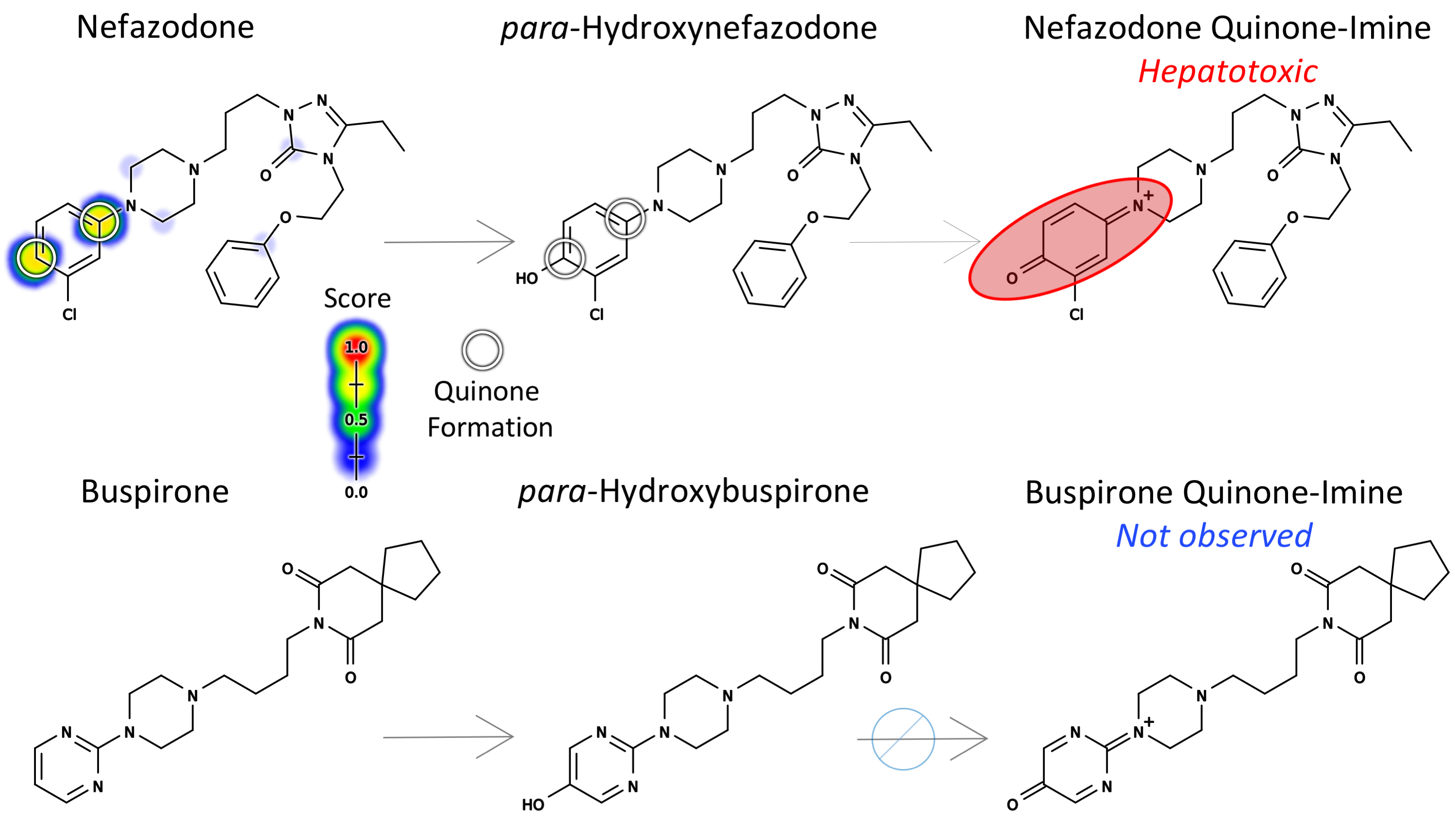
Abstract: Many adverse drug reactions are thought to be caused by electrophilically reactive drug metabolites that conjugate to nucleophilic sites within DNA and proteins,causing cancer or toxic immune responses. Quinone species, including quinone-imines, quinone-methides, and imine-methides, are electrophilic Michael acceptors that are often highly reactive, and comprise over 40% of all known reactive metabolites. Quinone metabolites are created by cytochromes P450 and peroxidases. For example, cytochromes P450 oxidize acetaminophen to N-acetyl-p-benzoquinone imine, which is electrophilically reactive and covalently binds to nucleophilic sites within proteins. This reactive quinone metabolite elicits a toxic immune response when acetaminophen exceeds a safe dose. Using a deep learning approach, this study reports the first published method for predicting quinone formation: the formation of a quinone species by metabolic oxidation. We model both one- and two-step quinone formation, enabling accurate quinone formation predictions in non-obvious cases. We predict atom pairs that form quinones with an AUC accuracy of 97.6%, and we identify molecules that form quinones with 88.2% AUC. By modeling the formation of quinones, one of the most common types of reactive metabolites, our method provides a rapid screening tool for a key drug toxicity risk. The XenoSite quinone formation model is available at https://swami.wustl.edu/xenosite/p/quinone.
Publication: http://pubs.acs.org/doi/abs/10.1021/acs.chemrestox.6b00385

You must log in to post a comment.