Chemical Computing Group Excellence Award
{{unknown}}
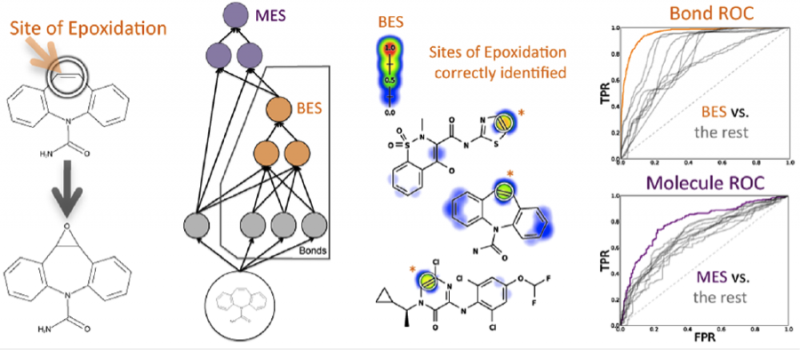 Drug toxicity is frequently caused by electrophilic reactive metabolites that covalently bind to proteins. Epoxides comprise … Continue Reading ››
Drug toxicity is frequently caused by electrophilic reactive metabolites that covalently bind to proteins. Epoxides comprise … Continue Reading ›› Managing missing measurements in small-molecule screens. Browning MR, Calhoun BT, Swamidass SJ. J Comput Aided Mol Des. 2013 … Continue Reading ››