Citation:
Hughes, Tyler B., Grover Paul Miller, and S. Joshua Swamidass. “Site of Reactivity Models Predict Molecular Reactivity of Diverse Chemicals with Glutathione.” Chemical Research in Toxicology (2015).
Abstract:
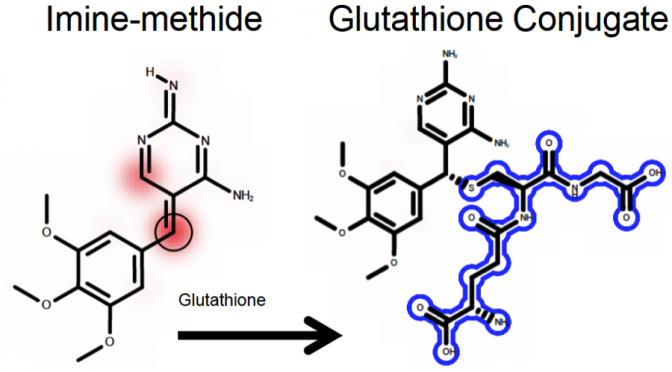
Drug toxicity is often caused by electrophilic reactive metabolites that covalently bind to proteins. Consequently, the quantitative strength of a molecule’s reactivity with glutathione (GSH) is a frequently used indicator of its toxicity. Through cysteine, GSH (and proteins) scavenges reactive molecules to form conjugates in the body. GSH conjugates to specific atoms in reactive molecules, their sites of reactivity. The value of knowing a molecule’s sites of reactivity is unexplored in the literature. This study tests the value of site of reactivity data that identifies the atoms within 1213 reactive molecules that conjugate to GSH and builds models to predict molecular reactivity with glutathione. An algorithm originally written to model sites of Cytochrome P450 metabolism (called XenoSite) finds clear patterns in molecular structure that identify sites of reactivity within reactive molecules with 90.8% accuracy and separate reactive and unreactive molecules with 80.6% accuracy. Furthermore, the model output strongly correlates with quantitative GSH reactivity data in chemically diverse, external datasets. Site of reactivity data is nearly unstudied in the literature prior to our efforts, yet contains a strong signal for reactivity that can be utilized to more accurately predict molecule reactivity and, eventually, toxicity.
Site: http://pubs.acs.org/doi/abs/10.1021/acs.chemrestox.5b00017

You must log in to post a comment.