Published online in Bioinformatics in February 2015
Extending P450 Site-of-Metabolism Models with Region-Resolution Data
Jed Zaretzki, Michael Browning, Tyler B. Hughes, S. Joshua Swamidass
Motivation: Cytochrome P450s are a family of enzymes responsible for the metabolism of approximately 90% of FDA approved drugs. Medicinal chemists often want to know which atoms of a … Continue Reading ›› Interviewed on “People Behind the Science”
Combined Analysis of Phenotypic and Target-Based Screening in Assay Networks
Citation:
Swamidass, S. J., Schillebeeckx, C. N., Matlock, M., Hurle, M. R., & Agarwal, P. (2014). Combined Analysis of Phenotypic and Target-Based Screening in Assay Networks. Journal of biomolecular screening, 1087057114523068.
Abstract:
Small-molecule screens are an integral part of drug discovery. Public domain data in PubChem alone represent more than
158 million measurements, 1.2 million molecules, and 4300 assays. … Continue Reading ››
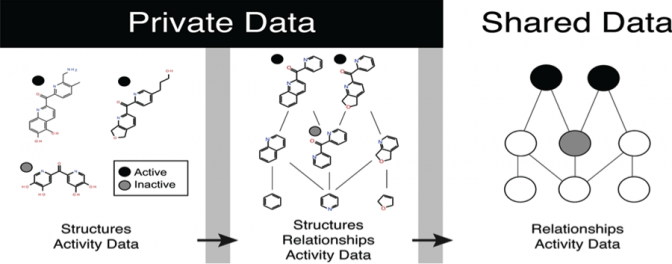
Sharing Chemical Relationships Does Not Reveal Structures
Citation:
Matlock, M., & Swamidass, S. J. (2013). Sharing Chemical Relationships Does Not Reveal Structures. Journal of chemical information and modeling, 54(1), 37-48.
Abstract: In this study, we propose a new, secure method of sharing useful chemical information from small-molecule libraries, without revealing the structures of the libraries’ molecules. Our method shares the relationship between molecules rather … Continue Reading ››
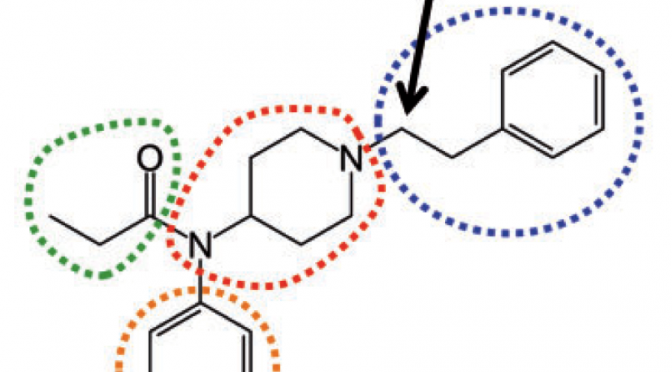
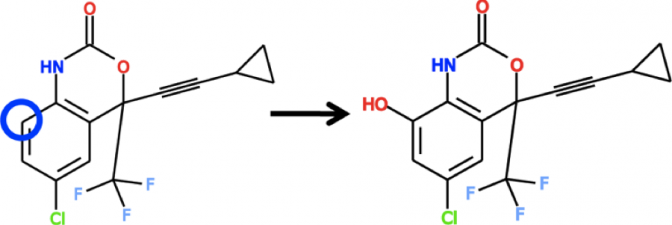
XenoSite: Accurately Predicting CYP-mediated Sites of Metabolism with Neural Networks
Web Server:
https://swami.wustl.edu/xenosite
Citation:
Zaretzki, J., Matlock, M., & Swamidass, S. J. (2013). XenoSite: Accurately predicting CYP-mediated sites of metabolism with neural networks. Journal of chemical information and modeling, 53(12), 3373-3383..
Abstract:
Understanding how xenobiotic molecules are metabolized is important because it influences the safety, efficacy, and dose of medicines and how they can be modified to improve these … Continue Reading ››
Round up of Recently Published Papers
Over the last several months we have published several papers. In the coming weeks, I will be posting abstracts and additional information for each of these papers. In the meantime, here are the citations of those that have hit print.
Managing missing measurements in small-molecule screens. Browning MR, Calhoun BT, Swamidass SJ. J Comput Aided Mol Des. 2013 … Continue Reading ››
World Drug Repositioning Conference
This week (December 4-5, 2012), I am honored to chair and present at the World Drug Repositioning Conference in Washington DC. This conference brings together several key industry groups to share insight into how they are discovering new uses for drugs in their pipelines. The second day of the conference, I will be presenting on technology developed … Continue Reading ››
New Post Doc: Jed Zaretzki
It is a pleasure to welcome our first postdoc to the Laboratory. Jed did his PhD under Curt Breneman, one of the leaders in Chemical Informatics, at the Rensselaer Exploratory Center for Cheminformatics Research, and his work was supported by Eli Lilly. His PhD focused on predicting how molecules are metabolized by P450 enzymes and culminated with the … Continue Reading ››
Understanding Screening Data with Workflows
The Current Group

Postdoctoral Researchers
- Jed Zaretzki
- Wee Kiang (starting in February 2013)
Scientific Programmers
- Michael Browning (Musician)
- Matthew Matlock
Past Members
- Brian Chen, PhD Student at Washington University (2011-2012, Programmer)
- Brad Calhoun, Software Programmer at Garmin (2010-2012, Programmer)
- Constantino Schillebeeckx, Photographer (2011-2012, Programmer)
- CJ Carey, PhD Student at University … Continue Reading ››